Working with scverse objects in backed mode#
In this tutorial we show how to work with scverse data objects without loading the full datasets in memory. AnnData and MuData objects are saved to disk as .h5ad and .h5mu files. These formats save HDF5 stores (via h5py) that can be accessed without loading heavy data matrices in memory (i.e. in backed mode), as a “read-only” object.
In this tutorial, we demonstrate how to work in backed mode for:
quickly inspecting the content of an h5ad/h5mu file
exploratory data analysis (e.g. plotting embeddings)
selecting a subset of interest in a very large dataset
from warnings import filterwarnings
# for datasets
import mudatasets
import muon as mu
import pandas as pd
import scanpy as sc
filterwarnings("ignore", category=FutureWarning)
filterwarnings("ignore", category=pd.errors.SettingWithCopyWarning)
sc.set_figure_params(fontsize=18, figsize=[6, 6])
Working with backed AnnData objects#
Let’s load an anndata object in memory
adata = sc.datasets.pbmc3k_processed()
adata = adata.raw.to_adata() # Store raw counts in adata.X
Let’s check the size of this AnnData object (i.e. the amount of RAM needed to load it)
# Convenience method for computing the size of objects
def print_size_in_MB(x):
print(f"{x.__sizeof__() / 1e6:.3} MB")
print_size_in_MB(adata)
21.2 MB
Now let’s save this object in .h5ad format and re-load it in backed (on-disk) mode.
adata.write_h5ad("./data/pbmc3k_processed_raw.h5ad")
adata = sc.read_h5ad("./data/pbmc3k_processed_raw.h5ad", backed=True)
Now the size of our AnnData object is about 3 MB, with the gene expression matrix saved only on disk
print_size_in_MB(adata)
3.28 MB
We can check whether an anndata object is loaded in backed mode using the adata.isbacked attribute
adata.isbacked
True
while the path to the file on disk is stored in adata.filename
adata.filename
PosixPath('data/pbmc3k_processed_raw.h5ad')
Exploratory data analysis#
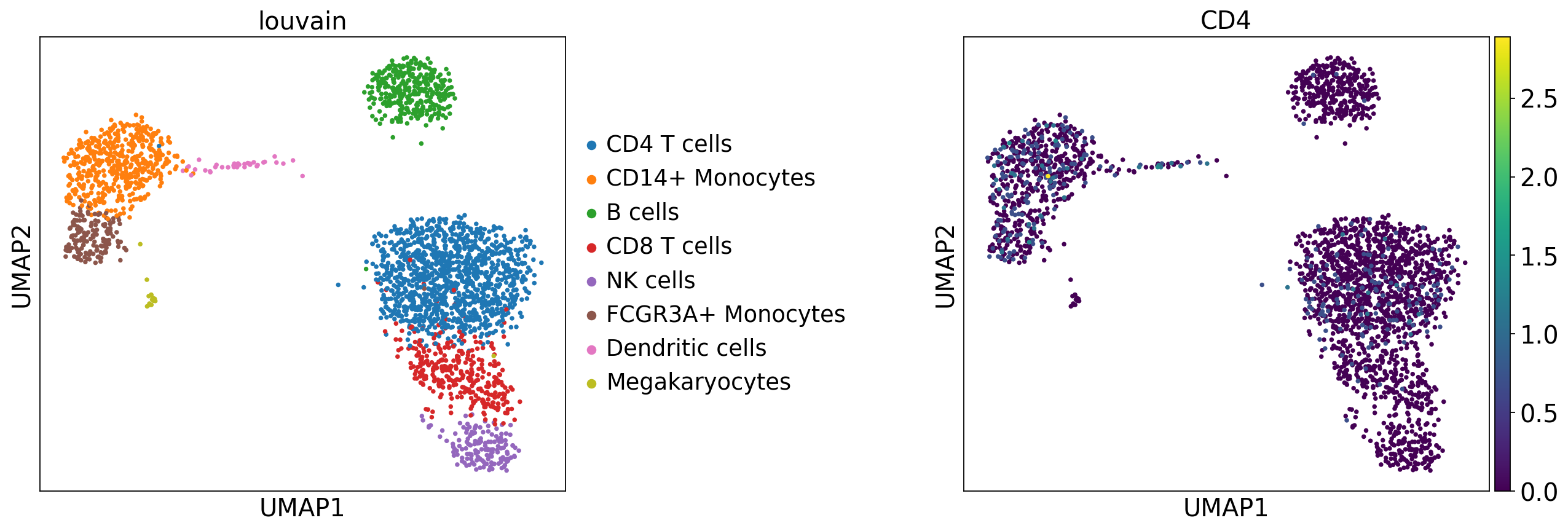
When loading an h5ad file in backed mode, we can use all scverse functions that require to read the data without having in memory the full adata.X matrix. For example we can plot embeddings or gene expression matrices. The gene expression values for a given gene or set of genes are loaded from disk on the function call.
The full gene expression matrix in adata.X is not loaded, but all the cell- and gene-level metadata is in memory and can be modified. Therefore we can run methods that modify obs/obsm/obsp. For example, we can compute KNN graphs and nearest neighbor embeddings
sc.pp.neighbors(adata, n_neighbors=20)
sc.tl.umap(adata)
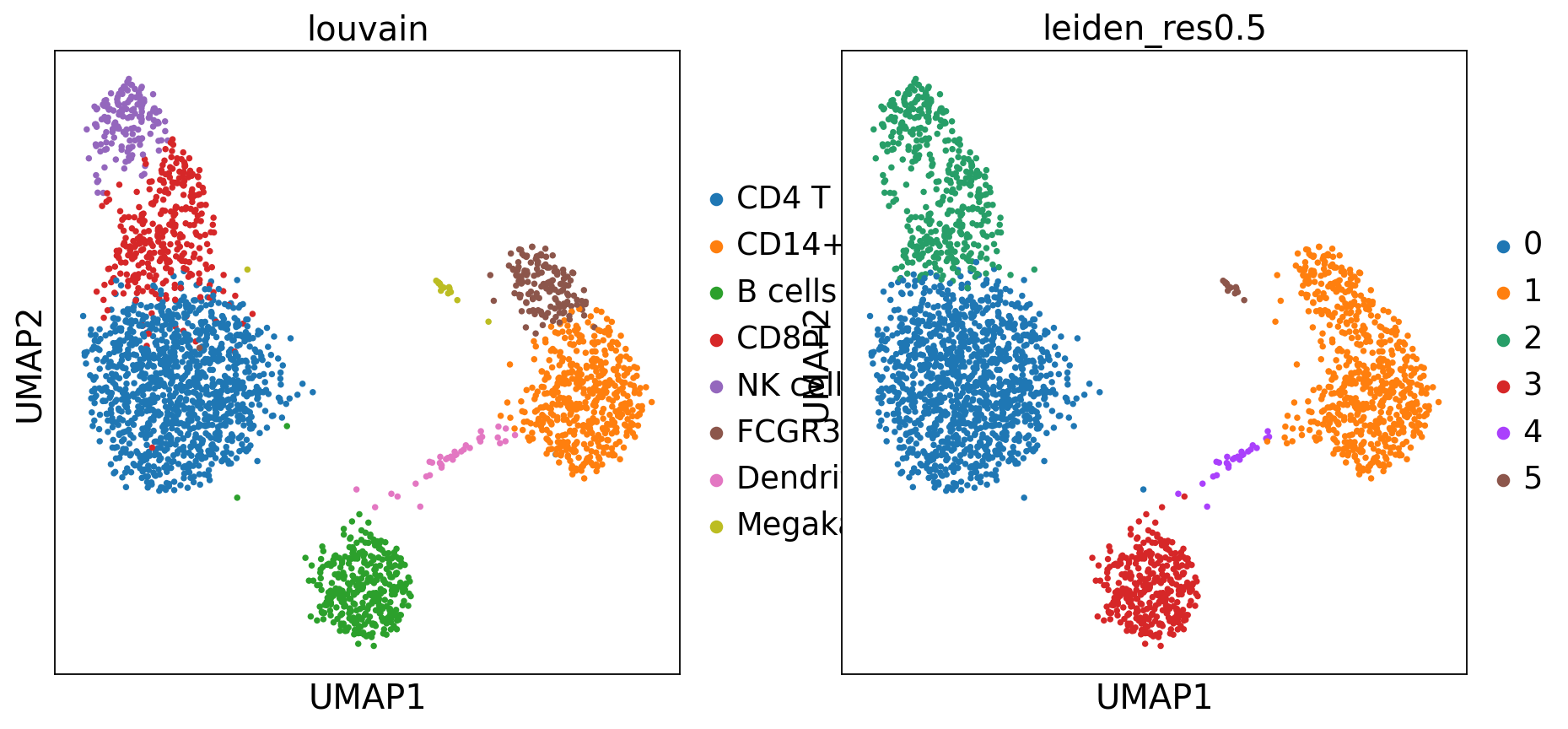
Or we can perform clustering
sc.tl.leiden(adata, resolution=0.5, key_added="leiden_res0.5")
adata.obs
| n_genes | percent_mito | n_counts | louvain | leiden_res0.5 | |
|---|---|---|---|---|---|
| index | |||||
| AAACATACAACCAC-1 | 781 | 0.030178 | 2419.0 | CD4 T cells | 0 |
| AAACATTGAGCTAC-1 | 1352 | 0.037936 | 4903.0 | B cells | 3 |
| AAACATTGATCAGC-1 | 1131 | 0.008897 | 3147.0 | CD4 T cells | 0 |
| AAACCGTGCTTCCG-1 | 960 | 0.017431 | 2639.0 | CD14+ Monocytes | 1 |
| AAACCGTGTATGCG-1 | 522 | 0.012245 | 980.0 | NK cells | 2 |
| ... | ... | ... | ... | ... | ... |
| TTTCGAACTCTCAT-1 | 1155 | 0.021104 | 3459.0 | CD14+ Monocytes | 1 |
| TTTCTACTGAGGCA-1 | 1227 | 0.009294 | 3443.0 | B cells | 3 |
| TTTCTACTTCCTCG-1 | 622 | 0.021971 | 1684.0 | B cells | 3 |
| TTTGCATGAGAGGC-1 | 454 | 0.020548 | 1022.0 | B cells | 3 |
| TTTGCATGCCTCAC-1 | 724 | 0.008065 | 1984.0 | CD4 T cells | 0 |
2638 rows × 5 columns
Extracting a subset of AnnData#
Another common use-case for backed mode is to inspect a dataset to then select and save a subset of data that we are interested in using for analysis.
For example, if we are interested in performing analysis just on T cells, we can subset the AnnData object and load in memory just the gene expression matrix of the subset of interest
T_clusters = ["CD4 T cells", "CD8 T cells"]
adata_subset = adata[adata.obs["louvain"].isin(T_clusters)].to_memory()
adata_subset
AnnData object with n_obs × n_vars = 1460 × 13714
obs: 'n_genes', 'percent_mito', 'n_counts', 'louvain', 'leiden_res0.5'
var: 'n_cells'
uns: 'draw_graph', 'louvain', 'louvain_colors', 'neighbors', 'pca', 'rank_genes_groups', 'umap', 'leiden', 'leiden_res0.5_colors'
obsm: 'X_draw_graph_fr', 'X_pca', 'X_tsne', 'X_umap'
obsp: 'connectivities', 'distances'
adata_subset.isbacked
False
If we want to keep working in backed mode with this data subset, we can copy it specifying a new .h5ad file on disk where this will be saved.
adata_subset = adata[adata.obs["louvain"].isin(T_clusters)].copy(filename="./data/pbmc3k_processed_CD4Tcells.h5ad")
adata_subset
AnnData object with n_obs × n_vars = 1460 × 13714 backed at 'data/pbmc3k_processed_CD4Tcells.h5ad'
obs: 'n_genes', 'percent_mito', 'n_counts', 'louvain', 'leiden_res0.5'
var: 'n_cells'
uns: 'draw_graph', 'leiden', 'leiden_res0.5_colors', 'louvain', 'louvain_colors', 'neighbors', 'pca', 'rank_genes_groups', 'umap'
obsm: 'X_draw_graph_fr', 'X_pca', 'X_tsne', 'X_umap'
obsp: 'connectivities', 'distances'
adata_subset.isbacked
True
Things you can’t do with AnnData in backed mode#
Backed mode loads gene expression matrices in “read-only” mode, so functions that attempt to modify adata.X will throw errors.
For example, normalizing with
sc.pp.normalize_total(adata)
Throws the following error
AttributeError: 'SparseDataset' object has no attribute 'sum'
but the gene expression matrix can be accessed directly using numerical indices.
adata.X[:, 0 : adata.n_vars]
<2638x13714 sparse matrix of type '<class 'numpy.float32'>'
with 2238732 stored elements in Compressed Sparse Row format>
Of note, in the current version of anndata the matrices in adata.layers are not backed.
Another caveat is that an object can be opened in backed mode only from one process/notebook simultaneously. Attempting to load it (with sc.read_h5ad) will throw an Input/Output error.
Save changes made to backed AnnData#
We can save the changes we’ve made to the backed anndata by writing back to the linked h5ad file.
adata.write_h5ad()
If we re-load this object from file, we will find all the updated information in the obs slot
adata = sc.read_h5ad("./data/pbmc3k_processed_raw.h5ad", backed=True)
adata
AnnData object with n_obs × n_vars = 2638 × 13714 backed at 'data/pbmc3k_processed_raw.h5ad'
obs: 'n_genes', 'percent_mito', 'n_counts', 'louvain', 'leiden_res0.5'
var: 'n_cells'
uns: 'draw_graph', 'leiden', 'leiden_res0.5_colors', 'louvain', 'louvain_colors', 'neighbors', 'pca', 'rank_genes_groups', 'umap'
obsm: 'X_draw_graph_fr', 'X_pca', 'X_tsne', 'X_umap'
obsp: 'connectivities', 'distances'
We can also save to a different file
adata.write_h5ad("./data/pbmc3k_processed_newclustering.h5ad")
Working with backed MuData objects#
Working with data on disk is particularly useful when working with data modalities different from gene expression, where the number of measured features is typically larger. For example, in data from scATAC-seq experiments accessibility is usually measured at hundreds of thousands of genomic regions, making feature x cell matrices prohibitively large to load in memory.
For this application, also MuData objects support working in backed mode.
# Download mudata object and save in h5mu format
mdata = mudatasets.load("pbmc10k_multiome")
mdata["rna"].var_names_make_unique()
mdata.write_h5mu("./data/pbmc10k_multiome.h5mu")
■ Loading filtered_feature_bc_matrix.h5...
Added `interval` annotation for features from /home/runner/mudatasets/pbmc10k_multiome/filtered_feature_bc_matrix.h5
We can see that the slot storing the scATAC accessibility matrix is quite large when loaded in memory
mdata["atac"].isbacked
False
print_size_in_MB(mdata["atac"])
7.16e+02 MB
Let’s now load the .h5mu object in backed mode using the read_h5mu function from the muon package.
mdata = mu.read_h5mu("./data/pbmc10k_multiome.h5mu", backed=True)
mdata
MuData object with n_obs × n_vars = 11909 × 144978 backed at 'data/pbmc10k_multiome.h5mu'
var: 'gene_ids', 'feature_types', 'genome', 'interval'
2 modalities
rna: 11909 x 36601
var: 'gene_ids', 'feature_types', 'genome', 'interval'
atac: 11909 x 108377
var: 'gene_ids', 'feature_types', 'genome', 'interval'Now the feature x cell matrices for both modalities are on-disk, and the object takes up a lot less memory.
print_size_in_MB(mdata["atac"])
31.6 MB
Working with only one modality in memory#
If we are analysing a multi-modal dataset, we might need to handle and process only one modality at a time. In this case we can keep the other modality on disk to reduce memory usage. For example, we can load the RNA modality only, to run clustering.
mdata.mod["rna"] = mdata.mod["rna"].to_memory()
mdata["rna"].isbacked
False
mdata["atac"].isbacked
True
Now we can run preprocessing of the expression data
rna = mdata.mod["rna"]
## Normalization
sc.pp.normalize_total(rna, target_sum=1e4)
sc.pp.log1p(rna)
## HVG selection
sc.pp.highly_variable_genes(rna, min_mean=0.02, max_mean=4, min_disp=0.5)
## Dim reduction and embeddings
sc.tl.pca(rna, svd_solver="arpack")
sc.pp.neighbors(rna, n_neighbors=10, n_pcs=20)
sc.tl.umap(rna)
sc.tl.leiden(rna, resolution=1.0)
Now all the changes are saved in the MuData object.
mdata["rna"]
AnnData object with n_obs × n_vars = 11909 × 36601
obs: 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'interval', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'log1p', 'hvg', 'pca', 'neighbors', 'umap', 'leiden', 'leiden_colors'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
obsp: 'distances', 'connectivities'
We can save changes to the h5mu file on disk, without needing to load the ATAC modality dataset
## Save log-normalized files in layers
# (write_h5mu doesn't rewrite .X if the file is in backed mode)
mdata["rna"].layers["lognorm_counts"] = mdata["rna"].X.copy()
mdata.write_h5mu()
Now we can reload the updated object to have both modalities on disk
mdata = mu.read_h5mu("./data/pbmc10k_multiome.h5mu", backed=True)
mdata
MuData object with n_obs × n_vars = 11909 × 144978 backed at 'data/pbmc10k_multiome.h5mu'
var: 'gene_ids', 'feature_types', 'genome', 'interval'
2 modalities
rna: 11909 x 36601
obs: 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'interval', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'hvg', 'leiden', 'leiden_colors', 'log1p', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'lognorm_counts'
obsp: 'connectivities', 'distances'
atac: 11909 x 108377
var: 'gene_ids', 'feature_types', 'genome', 'interval'mdata["atac"].isbacked and mdata["rna"].isbacked
True
Extracting a MuData subset#
Similarly to the AnnData case, we can use the backed mode to identify a subset of data that we are interested in, and save it as a smaller object to load into memory.
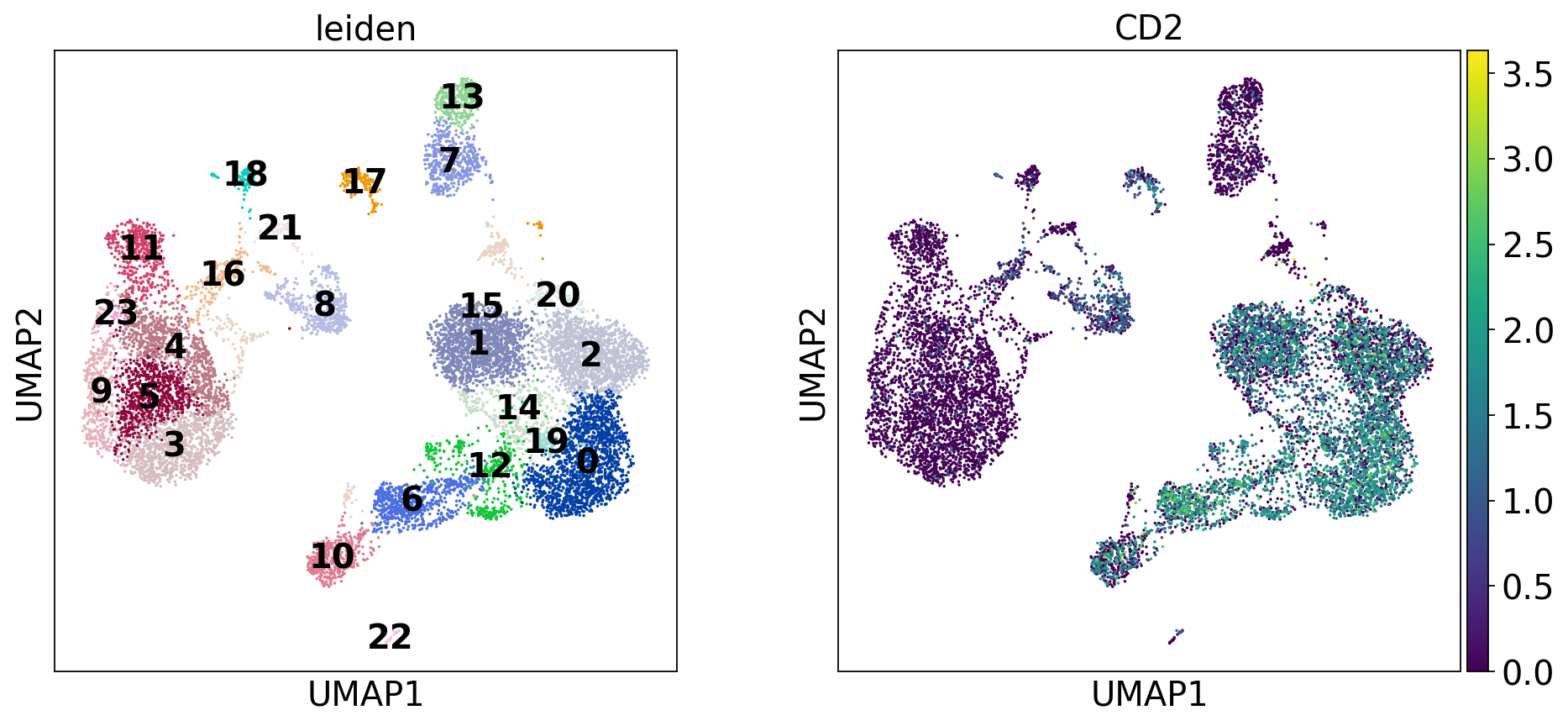
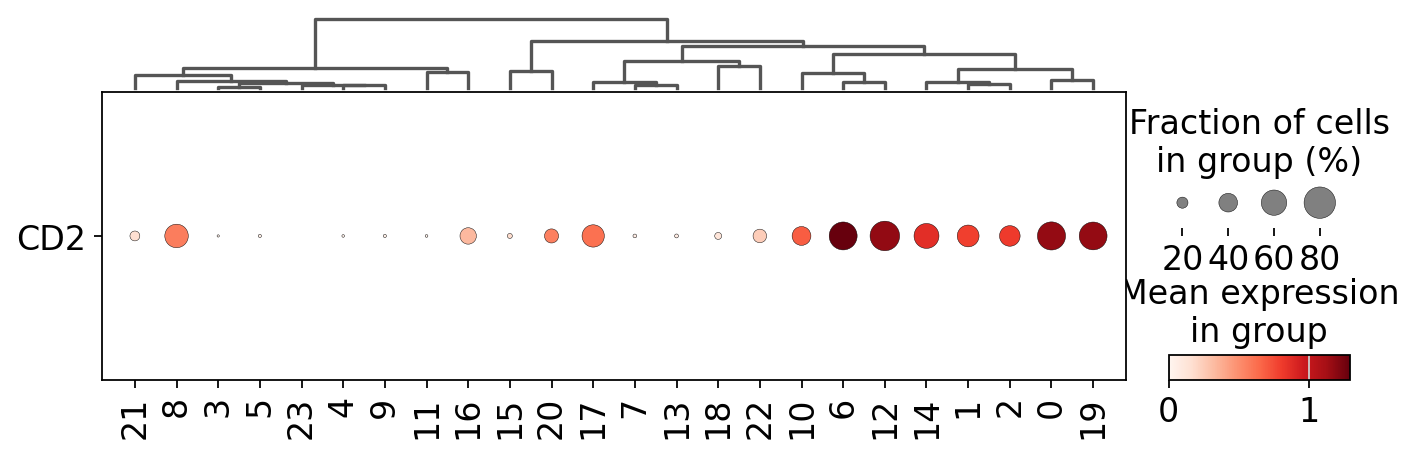
For example, we could subset the PBMC dataset to include only the T and NK cells (clusters expressing CD2)
sc.pl.dotplot(mdata["rna"], var_names=["CD2"], groupby="leiden", figsize=(10, 3), dendrogram=True, swap_axes=True)
nkt_cell_clusters = ["19", "20", "13", "11", "9", "14", "19", "5", "0", "1", "3"]
mdata[mdata.obs["rna:leiden"].isin(nkt_cell_clusters)]
View of MuData object with n_obs × n_vars = 6389 × 144978 backed at 'data/pbmc10k_multiome.h5mu'
var: 'gene_ids', 'feature_types', 'genome', 'interval'
2 modalities
rna: 6389 x 36601
obs: 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'interval', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'hvg', 'leiden', 'leiden_colors', 'log1p', 'neighbors', 'pca', 'umap', 'dendrogram_leiden'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'lognorm_counts'
obsp: 'connectivities', 'distances'
atac: 6389 x 108377
var: 'gene_ids', 'feature_types', 'genome', 'interval'We can save this data subset in a new .h5mu object using the copy method
mdata_nkt = mdata[mdata.obs["rna:leiden"].isin(nkt_cell_clusters)].copy(
filename="./data/pbmc10k_multiome.NKTcells.h5mu"
)
mdata_nkt
MuData object with n_obs × n_vars = 6389 × 144978 backed at 'data/pbmc10k_multiome.NKTcells.h5mu'
var: 'gene_ids', 'feature_types', 'genome', 'interval'
2 modalities
rna: 6389 x 36601
obs: 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'interval', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'dendrogram_leiden', 'hvg', 'leiden', 'leiden_colors', 'log1p', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'lognorm_counts'
obsp: 'connectivities', 'distances'
atac: 6389 x 108377
var: 'gene_ids', 'feature_types', 'genome', 'interval'We can load this object in memory by setting mdata.filename to None
mdata_nkt
MuData object with n_obs × n_vars = 6389 × 144978 backed at 'data/pbmc10k_multiome.NKTcells.h5mu'
var: 'gene_ids', 'feature_types', 'genome', 'interval'
2 modalities
rna: 6389 x 36601
obs: 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'interval', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'dendrogram_leiden', 'hvg', 'leiden', 'leiden_colors', 'log1p', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'lognorm_counts'
obsp: 'connectivities', 'distances'
atac: 6389 x 108377
var: 'gene_ids', 'feature_types', 'genome', 'interval'mdata_nkt.filename = None
mdata_nkt.isbacked
False
or reading from file
mdata_nkt = mu.read_h5mu("./data/pbmc10k_multiome.NKTcells.h5mu")
mdata_nkt
MuData object with n_obs × n_vars = 6389 × 144978
var: 'gene_ids', 'feature_types', 'genome', 'interval'
2 modalities
rna: 6389 x 36601
obs: 'leiden'
var: 'gene_ids', 'feature_types', 'genome', 'interval', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'dendrogram_leiden', 'hvg', 'leiden', 'leiden_colors', 'log1p', 'neighbors', 'pca', 'umap'
obsm: 'X_pca', 'X_umap'
varm: 'PCs'
layers: 'lognorm_counts'
obsp: 'connectivities', 'distances'
atac: 6389 x 108377
var: 'gene_ids', 'feature_types', 'genome', 'interval'mdata_nkt.isbacked
False
Things you can’t do with MuData in backed mode#
The current MuData implementation doesn’t support having a different number of obs between modalities in backed mode.